PhD defense
PhD defended on February 14, 2007 at INRIA Sophia Antipolis.
Reviewers:
- Patrick Clarysse, CREATIS CNRS, Lyon.
- Louis Collins, McConnell Brain Imaging Center, McGill University, Montreal.
Jury:
- Nicholas Ayache - President. INRIA Asclepios, Sophia Antipolis.
- Pierre-Yves Bondiau - Examiner. Cenre Antoine Lacassagne, Nice.
- Guido Gerig - Examiner. University of North Carolina.
- Vincent Grégoire - Examiner. Cliniques Universitaires Saint Luc, Université Catholique de Louvain.
- Grégoire Malandain - Advisor. INRIA Asclepios, Sophia Antipolis.
- Hanna Kafrouni - Invited. DOSIsoft S.A. Cachan
Documents:
- Thesis Manuscript (in french).
- PhD defense slides (in english).
For those interested, I have put online the LaTeX template (both for french and english PhDs) I wrote and used to write the PhD thesis at this address. Both are given with very short examples showing how to use these templates.
Atlas-based segmentation
Work with DOSIsoft SA, Cachan, INRIA Sophia Antipolis and Centre Antoine Lacassagne, Nice.
The treatment of cerebral tumors may involve surgery, radiotherapy, or chemotherapy. Thanks to recent technological advances (on-line definition of the shape of the irradiation beam, irradiation intensity modulation during the treatment), conformal radiotherapy allows a high precision irradiation (homogeneous dose distribution within complex shapes), permitting an improvement of local control and the reduction of the complications. In order to determine the best characteristics of the treatment planning, and to provide the patient follow-up, it is necessary to accurately locate the tumor and all the structures of interest in the brain. An automatic segmentation algorithm of all the critical structures in a patient image is then an invaluable tool for radiotherapy [3].
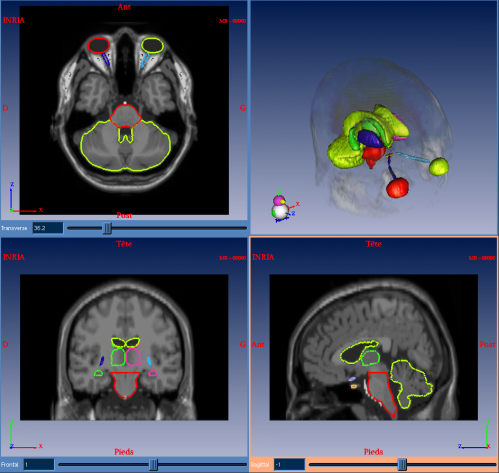
In order to extract all these structures in a specific patient's image, we chose to build a numerical reference atlas of all the structures we are interested in. This atlas is composed of a couple of images (see figure 1) : one simulated T1 MR image and its corresponding labelization.
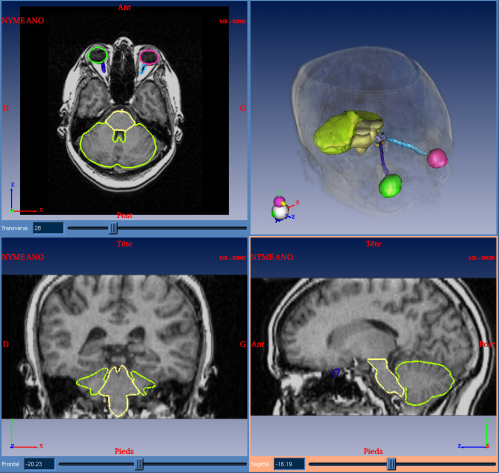
Registering a brain atlas on the patient's MRI for labeling brain structures is indeed an interesting alternative to manual segmentation. This technique consists in bringing the atlas MRI in global correspondence with the patient's image thanks to an affine transformation. Then the atlas image is warped locally to the patient thanks to an elastic registration. Applying the obtained transformations to the structures available in the label image allow us to segment automatically the patient's anatomy.
My thesis subject is then about the study and use of non-rigid registration algorithms in order to best register the patient image on the atlas. After this study, I have chosen to develop and use a registration algorithm, which is looking for a multi-affine transformation (see this page for more details and [1,2]). This algorithm allows to get smoother contours than with a non-rigid dense registration, while being as precise. Finally, the parameters can be fixed once for all the patients, even if the images are acquired in different centers. This algorithm has been integrated as a module in DOSISoft software for automatic segmentation of critical structures for radiotherapy planning (see figure 2).
|
Bibliography
-
An Efficient Locally Affine Framework for the Smooth Registration of Anatomical Structures
O. Commowick, V. Arsigny, A. Isambert, J. Costa, F. Dhermain, F. Bidault, P.-Y. Bondiau, N. Ayache and G. Malandain.
Medical Image Analysis, 12(4):427-441, August 2008.

-
An Efficient Locally Affine Framework for the Registration of Anatomical Structures
O. Commowick, V. Arsigny, J. Costa, N. Ayache, and G. Malandain.
In Proceedings of the Third IEEE International Symposium on Biomedical Imaging (ISBI 2006), pages 478-481, April 2006.
-
Atlas-based Automatic Segmentation of MR Images: Validation Study on the Brainstem in Radiotherapy
P.-Y. Bondiau, G. Malandain, S. Chanalet, P.-Y. Marcy, J.-L. Habrand, F. Fauchon, P. Paquis, A. Courdi, O. Commowick, I. Rutten, and N. Ayache.
Int J Radiat Oncol Biol Phys, 61(1):289-98, January 2005.