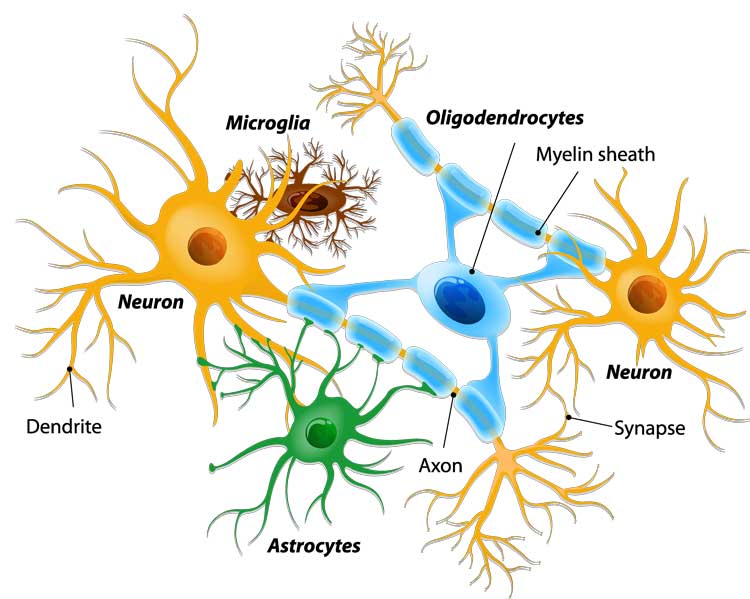
One of my major research interests lies in the modeling of the brain microstructure from diffusion MRI, and the estimation of these models from the acquired signals, often in clinical conditions. On these topics, I am particularly interested in trying to get an easily interpretable model of the brain microstructure i.e. able to represent the different tissues of the brain as illustrated in Fig. 1.
|
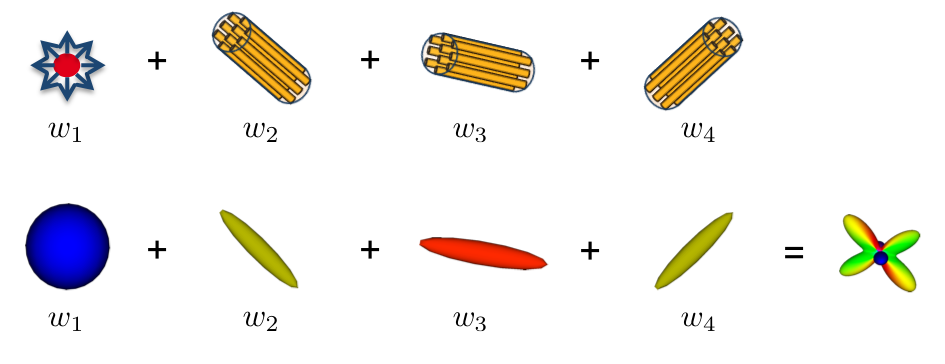
The most used model in the literature remains the tensor but it is limited to model properly crossing fibers. Moreover, the information in it is entangled as it comes from different tissues and is merged into just a single compartment. I study in my research models to go beyond this model: the multi-compartment models of diffusion or diffusion compartment models. These are very interesting as they model each tissue class or fiber bindle by a single compartment (see Fig. 2), thus providing a much more understandable model in the clinics.
|
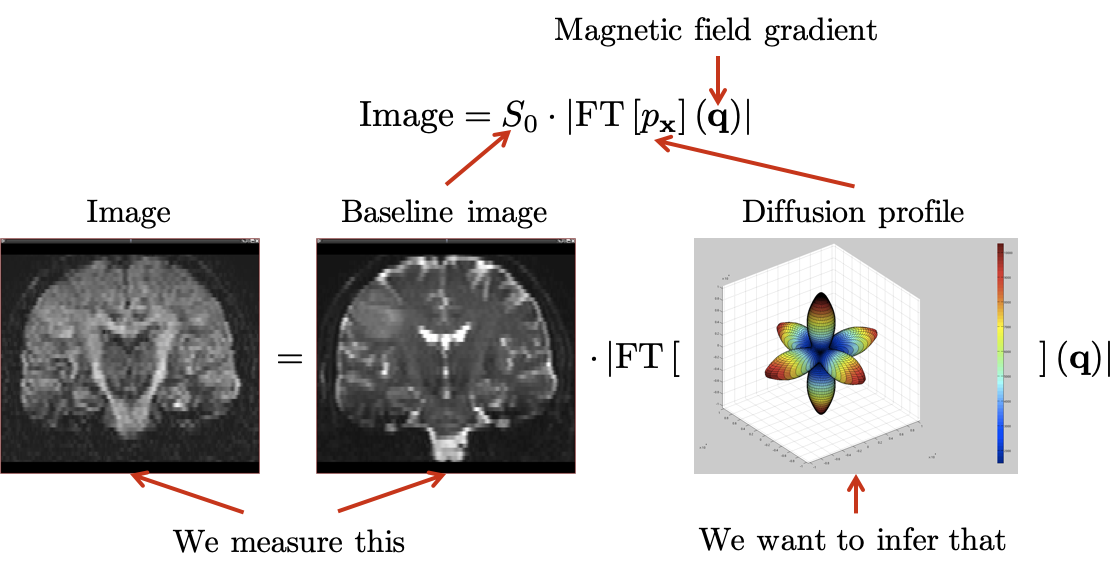
This model is however much more complex to estimate from the data acquired from the MRI scanner (as illustrated in Fig. 3), especially when dealing with clinical acquisitions. My research focuses on both aspects: estimating these models robustly, adapting models for clinical acquisitions, be as generic as possible to handle all possible compartments available in the literature.
|
|